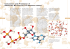P-class pumps
Transcription
P-class pumps
Active Transport ADP + Pi S2 S1 ATP Side 1 Side 2 Active transport enzymes couple net solute movement across a membrane to ATP hydrolysis. An active transport pump may be a uniporter or antiporter. ATP-dependent ion pumps are grouped into classes based on transport mechanism, as well as genetic & structural homology. Examples include: P-class pumps F-class (e.g., F1Fo-ATPase) & related V-class pumps. ABC (ATP binding cassette) transporters, which catalyze transmembrane movements of various organic compounds including amphipathic lipids and drugs (e.g. P-glycoprotein multidrug resistance) P-class ion pumps are a gene family exhibiting sequence homology. They include: Na+,K+-ATPase, in plasma membranes of most animal cells is an antiport pump. It catalyzes ATP-dependent transport of Na+ out of a cell in exchange for K+ entering. (H+, K+)-ATPase, involved in acid secretion in the stomach is an antiport pump. It catalyzes transport of H+ out of the gastric parietal cell (toward the stomach lumen) in exchange for K+ entering the cell. P-class pumps (cont): Ca++-ATPases, in endoplasmic reticulum (ER) and plasma membranes, catalyze ATPdependent transport of Ca++ away from the cytosol, into the ER lumen or out of the cell. Some evidence indicates that these pumps are antiporters, transporting protons in the opposite direction. Ca++-ATPase pumps function to keep cytosolic Ca++ low, allowing Ca++ to serve as a signal. The reaction mechanism for a P-class ion pump involves transient covalent modification of the enzyme. O Enzyme-C OH ATP Pi ADP H2O O Enzyme- C O O P O- O- P-Class Pumps At one stage of the reaction cycle, phosphate is transferred from ATP to the carboxyl of a Glu or Asp side-chain, forming a “high energy” anhydride linkage (~P). At a later stage in the reaction cycle, the Pi is released by hydrolysis. The Prominent Members in the Family of P-Type ATPases have different number of subunits tissue Ca-ATPase (SR) muscles subunits a AA 994 Na,K-ATPase ubiquitary ab(g) 1013/295 H,K-ATPase stomach ab 1033/291 General Post-Albers reaction scheme for P-type ATPases E~P-Ca++2 E~P-Ca++2 ADP The ER Ca++ pump is called SERCA: Sarco(Endo)plasmic Reticulum Ca++-ATPase. ATP 2Ca++ E-Ca++2 2Ca++ E Pi cytosol membrane In this diagram of SERCA reaction cycle, conformational changes altering accessibility of Ca++-binding sites to the cytosol or ER lumen are depicted as positional changes. ER lumen Reaction cycle: 1. 2 Ca++ bind tightly from the cytosolic side, stabilizing the conformation that allows ATP to react with an active site aspartate residue. E~P-Ca++2 E~P-Ca++2 ADP ATP 2Ca++ E-Ca++2 2Ca++ E Pi cytosol membrane ER lumen 2. Phosphorylation of the active site aspartate induces a conformational change that • shifts accessibility of the 2 Ca++ binding sites from one side of the membrane to the other, & • lowers the affinity of the binding sites for Ca++. E~P-Ca++2 E~P-Ca++2 ADP ATP 2Ca++ E-Ca++2 2Ca++ E Pi cytosol membrane ER lumen 3. Ca++ dissociates into the ER lumen. 4. Ca++ dissociation promotes • hydrolysis of Pi from the enzyme Asp • conformational change (recovery) that causes Ca++ binding sites to be accessible again from the cytosol. Asp351 This X-ray structure of muscle SERCA (Ca++ATPase) shows 2 Ca++ ions (colored magenta) bound between transmembrane a-helices in the membrane domain. cytosolic domain 2 Ca++ PDB 1EUL membrane domain Muscle SERCA Active site Asp351, which is transiently phosphorylated during catalysis, is located in a cytosolic domain, far from the Ca++ binding sites. SERCA structure has been determined in the presence & absence of Ca++, with or without substrate or product analogs and inhibitors. Substantial differences in conformation have been interpreted as corresponding to different stages of the reaction cycle. Large conformational changes in the cytosolic domain of SERCA are accompanied by deformation & changes in position & tilt of transmembrane a-helices. The data indicate that when Ca++ dissociates: • water molecules enter Ca++ binding sites • charge compensation is provided by protonation of Ca++-binding residues. ++ Ca SERCA Conformational Cycle enzyme phosphorylation phosphate hydrolysis This simplified cartoon represents the proposed variation in accessibility & affinity of Ca++-binding sites during the reaction cycle. Only 2 transmembrane a-helices are represented, and the cytosolic domain of SERCA is omitted. Ca-ATPase H,K-ATPase Na,K-ATPase Sequence comparison of the TM helices in Ptype ATPases of various subfamilies conserved residues as compared to the ATP2A1 Ca2+ binding site sequence non-conserved residues Topology and architecture of the catalytic subunits of P-type ATPases. a strikingly similar fold despite strong sequence divergence N-domain binds ATP and serves as a built-in protein kinase, which autophosphorylates the P-domain. The A-domain acts as an intrinsic protein phosphatase dephosphorylating the P-domain later in the catalytic cycle. The process of phosphorylation and dephosphorylation is tightly coupled to formation and deformation of high-affinity transport-binding sites in the M domain by an allosteric mechanism P-type ATPases involved in neuronal disorders E1WCa2+ and E2(TG) forms of Ca2+ -ATPase in lipid bilayer Toyoshima, Nomura, Sugita, FEBS Letters 555 (2003) 106-110 Toyoshima, Nomura, Sugita, FEBS Letters 555 (2003) 106-110 Rearrangements of the transmembrane helices between E1WCa (violet) and E2(TG) (light green) forms Toyoshima, Nomura, Sugita, FEBS Letters 555 (2003) 106-110 MacLennan/Green, NATURE, Vol 405, 2000, 633-634 Overview of the structure of the Ca 2+-ATPase Models of E1 and E2 forms of the a-subunit of Na,K-ATPase based on the high-resolution structure of Ca-ATPase (1EUL) Annu. Rev. Physiol. 2003 .65 : 817–49 Pump Function: Post-Albers Cycle Apell, J. Rev Physiol Biochem Pharmacol (2003) 150:1–35 Pump Function: Post-Albers Cycle Ping-Pong Mechanism backward Apell, J. Rev Physiol Biochem Pharmacol (2003) 150:1–35 Pump Function: Post-Albers Cycle Ping-Pong Mechanism Stoichiometry Na,K-ATPase: 3 Na+ / 2 K+ H,K-ATPase: 2 H + / 2 K+ SR Ca-ATPase: 2 Ca2+ / 2 H+ Apell, J. Rev Physiol Biochem Pharmacol (2003) 150:1–35 Pump Function: Post-Albers Cycle Ping-Pong Mechanism Stoichiometry Na,K-ATPase: 3 Na+ / 2 K+ H,K-ATPase: 2 H+ / 2 K+ SR Ca-ATPase: 2 Ca2+ / 2 H+ Electrogenicity Na,K-ATPase: H,K-ATPase: SR Ca-ATPase: +1 0 +2 Apell, J. Rev Physiol Biochem Pharmacol (2003) 150:1–35 The Channel Concept of the Ion Pumps Apell, J. Rev Physiol Biochem Pharmacol (2003) 150:1–35 Experimental Methods Charge movements by electrophysiological methods by fluorescence methods RH421 Method RH421 Standard Experiment: reveals electrogenic partial reaction Apell, Bioelectrochemistry 63 (2004) 149– 156 In the E1 conformation of the Na,K-ATPase the two binding sites which bind K+ or Na+ ions are always occupied in physiological pH, if not by transported cations then by H+ ions. The fluorescence decrease with increasing ion concentrations indicates import of positive charge into the membrane domain of the ATPases Apell, Bioelectrochemistry 63 (2004) 149– 156 Hypothetical Model of Cytoplasmic Na+ Binding Apell, J. Membrane Biol. 180, 1–9 (2001) Hypothetical Model CytoplasmicNa Na++ Binding Binding Hypothetical Model ofof Cytoplasmic Apell, J. Membrane Biol. 180, 1–9 (2001) 2 Hypothetical Model of Cytoplasmic Na+ Binding Hypothetical Model of Cytoplasmic Na+ Binding Apell, J. Membrane Biol. 180, 1–9 (2001) 3 Methods of Investigation Charge Movements by electrophysiological techniques Experimental Methods Charge movements by electrophysiological methods by fluorescence methods Conformational changes by fluorescence methods FITC Methode pH and conformation dependence of the 5-IAF label covalently linked to rabbit a1 Na,KATPase. Titration in different conformations explains the F shift during pump cycle at constant bulk pH ATP-dependent Ca2+ uptake, H+ counter transport, and development of transmembrane electrical potential at low temperature. oxonol VI Pyranine (HPTS) arsenazo III [Yu et al. 1994] Energetics of selected reaction steps from rabbit kidney Na,K-ATPase