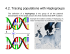
Population structure and identification of two matrilinear and one
Transcription
Population structure and identification of two matrilinear and one
Estuarine, Coastal and Shelf Science xxx (2014) 1e10 Contents lists available at ScienceDirect Estuarine, Coastal and Shelf Science journal homepage: www.elsevier.com/locate/ecss Population structure and identification of two matrilinear and one patrilinear mitochondrial lineages in the mussel Mytella charruana Thainara Oliveira de Souza a, 1, Francisco Arimateia dos Santos Alves a, b, 1, Colin Robert Beasley c, Luiz Ricardo Lopes de Simone d, Nelane do Socorro Marques-Silva a, Guilherme da Cruz Santos-Neto a, e, Claudia Helena Tagliaro a, * , Campus de Bragança, Instituto de Estudos Costeiros, Laborato ~o e Biologia Evolutiva, Alameda Leandro rio de Conservaça Universidade Federal do Para Ribeiro, s/n, Bragança, PA, CEP 68600-000, Brazil b ~o de Meio Ambiente, Laborato ^ndia, Ananindeua, PA, CEP rio de Biologia Ambiental, Rodovia BR 316, Km 07, s/n, Levila Instituto Evandro Chagas, Seça 67030-070, Brazil c , Campus de Bragança, Instituto de Estudos Costeiros, Laborato rio de Moluscos, Alameda Leandro Ribeiro, s/n, Bragança, PA, Universidade Federal do Para CEP 68600-000, Brazil d ~o Paulo, Caixa Postal 42494, Sa ~o Paulo, SP, CEP 04299-970, Brazil Museu de Zoologia da Universidade de Sa e , Campus de Abaetetuba, Rua Rio de Janeiro, 3322, Abaetetuba, PA, CEP 68440-000, Brazil Instituto Federal de Ci^ encia e Tecnologia do Para a a r t i c l e i n f o a b s t r a c t Article history: Accepted 10 November 2014 Available online xxx The mitochondrial gene cytochrome c oxidase subunit I (COI) was sequenced from Mytella charruana (N ¼ 243) at 10 Brazilian coastal localities to search for cryptic species, doubly uniparental inheritance and investigate genetic population structure and demography. Three haplogroups were found: two matrilinear (A and B) in males and females, and one patrilinear (C) found only in males. The p-distances were 0.0624 (A and B), 0.2097 (A and C) and 0.2081 (B and C). Coalescence of M. charruana occurred around 12.5 Mya, and the origins of the lineages were 3.4 and 4 Mya (matrilinear A and B) and 51.2 Mya (patrilinear), which split before the separation of the genera Perna and Mytella. All individuals from the northern coast of Brazil belonged to haplogroup A, whereas haplogroup B predominated among individuals from the eastern and northeastern coasts, with one exception, Goiana. Haplogroup C was found in males from the northern to the eastern coast. GenBank sequences of M. charruana from Colombia, Ecuador and four populations introduced to the USA joined Brazilian haplogroup B. Nuclear gene 18SITS1 sequences confirmed that all specimens belong to the same species. Four populations from the northern coast of Brazil were homogenous with evidence of recent population expansion. All populations from the northeastern and eastern coasts of Brazil were significantly structured (pairwise FST and AMOVA). The heterogeneity among Brazilian populations requires that relocation for aquaculture be preceded by genetic identification of the haplogroups. Differences in salinity and temperature may have selected for distinct lineages of mussels and changing conditions in coasts and estuaries may allow only resistant lineages of mussel to persist with the loss of others. In the light of global climate change, more detailed data on temperature, pH, salinity and local currents could help explain the genetic structuring observed among populations of Brazilian M. charruana. © 2014 Elsevier Ltd. All rights reserved. Keywords: genetic diversity geographic isolation population genetics DUI Mytilidae 1. Introduction Mytella charruana (d'Orbigny, 1842) (Mytilidae, Bivalvia), the charru mussel, is native to Central and South America where it * Corresponding author. E-mail address: [email protected] (C.H. Tagliaro). 1 Both authors contributed equally to this research. ranges, along the Pacific coast, from Guayamas, Mexico to southern Ecuador and the Galapagos Islands (Rios, 1994; Cardenas and Aranda, 2000) and, along the Atlantic coast, from Colombia to Argentina (Keen, 1971). Introíni et al. (2010) listed the diverse synonyms attributed to M. charruana, including Mytella falcata d'Orbigny, 1846, Modiola falcata Von Ihering, 1897, Mytilus strigatus Von Ihering, 1900, Mytilus arciformis Dall, 1909, Mytilus mundahuensis Duarte, 1926 and Modiolus falcatus Morretes, 1949. http://dx.doi.org/10.1016/j.ecss.2014.11.009 0272-7714/© 2014 Elsevier Ltd. All rights reserved. Please cite this article in press as: de Souza, T.O., et al., Population structure and identification of two matrilinear and one patrilinear mitochondrial lineages in the mussel Mytella charruana, Estuarine, Coastal and Shelf Science (2014), http://dx.doi.org/10.1016/j.ecss.2014.11.009 2 T.O. de Souza et al. / Estuarine, Coastal and Shelf Science xxx (2014) 1e10 Laboratory experiments show that Mytella charruana (synonym of Mytella falcata) planktonic larvae begin to settle on the substrate and Carvalheira, 1972), only after at least seven days (Paranagua thus allowing a high level of dispersion. The latter is common in species with a long planktonic larval stage and results in greater gene flow and low levels of population genetic structure (Hoskin, 1997; Murray-Jones and Ayre, 1997; Collin, 2001), in contrast to species with direct development (Collin, 2001). Environmental factors, such as temperature, salinity, physical barriers, long geographic distances, and oceanic current patterns, may, however, limit gene flow in species with planktonic development (Launey et al., 2002). The charru mussel is an euryhaline species (Leonel and Silva, 1988; Gillis et al., 2009) that tolerates salinities varying between 2 and 40 (Yuan et al., 2010). These bivalves may be found in shallow coastal lagoons and in muddy areas of bays and estuaries (Sibaja, 1988) in water temperatures of between 6 C and 31 C (Brodsky et al., 2009). Along the eastern coast of the United States, the charru mussel is considered an invasive species and has been found in Florida since 1986, most likely being introduced via ballast water or encrusted on ship hulls (Gillis et al., 2009). Molecular studies carried out on marine and freshwater bivalves show evidence for the presence of two lineages of mitochondrial DNA (mtDNA), one maternal (mtDNA F) and another paternal (mtDNA M) in origin due to doubly uniparental inheritance (DUI) of mtDNA (Skibinski et al., 1994; Hoeh et al., 1996; Theologidis et al., 2008). In this pattern of inheritance, females are typically homoplasmic for mtDNA F and transmit it to both sons and daughters, whereas males are heteroplasmic, carrying both mitochondrial genomes, whereas mtDNA F is predominant in somatic cells and M in gonadal cells, but transmitting mtDNA M only to their sons (Hoeh et al., 1996, 2002; Zouros, 2013). Although both lineages evolve independently (Hoeh et al., 1996; Breton et al., 2007), mtDNA M evolution is more rapid (Hoeh et al., 1997; Passamonti and Ghiselli, 2009). DUI has been widely studied in bivalves and has been recorded in the Mytilidae (Breton et al., 2007; Passamonti and Ghiselli, 2009; Zouros, 2013). Alves et al. (2012) detected mitochondrial heteroplasmy due to DUI in Mytella charruana from the Brazilian coast, showing evidence for the presence of this type of inheritance in yet another species of mytilid. According to the latter authors, intraspecific divergence between mtDNA F and M in M. charruana cytochrome c oxidase subunit I (COI) varied between 20.5 and 20.8%. However, the DUI pattern of inheritance was not found in three species of the mussel Perna (Wood et al., 2007) and, according to Gillis et al. (2009), DUI appeared to be absent in M. guyanensis and M. charruana. The discrepancy between the results of the latter two studies regarding M. charruana is explained by Alves et al. (2012) as a consequence of the choice of primers used by Gillis et al. (2009), that were only able to amplify COI fragments of matrilinear lineages. An ancient polymorphism (L and M groups) with a 7.3% COI sequence divergence was found between populations of the mussel Brachidontes pharaonis in the Mediterranean Sea and in the Red Sea between which, despite differences in haplotype frequencies, gene flow appears to be extensive (Sirna-Terranova et al., 2006). However, the population from Salina di Marsala had private haplotypes from L and M groups in high frequencies and the researchers hypothesize that these haplotypes are selectively advantageous in waters with high temperature and salinity. A more widespread analysis of the genus Brachidontes by the same authors, using COI and 16S rDNA mitochondrial sequences, revealed three geographically distinct monophyletic clades: B. pharonis from the Mediterranean and Red Seas, B. variabilis from the Indian Ocean that also has DUI, and B. variabilis from the western Pacific Ocean, which have divergence values corresponding to interspecific values in other bivalves and thus belong to three cryptic species (SirnaTerranova et al., 2007). Oliveira et al. (2005), using allozymes, found intraspecific structure among populations of Mytella charruana and M. guyanensis from the Brazilian coast, and, although analyzes indicated limited gene flow, the observed structure was not according to the isolation by geographic distance model. These authors were unable to give reasons for the retention of gene flow due to the lack of detailed studies of larval dispersion and settlement. On the other hand, Gillis et al. (2009) analyzing molecular data (COI) of two invasive populations of M. charruana in Florida (USA) and two native populations from South America (Cartagena in Colombia and Guayaquil in Ecuador), found distinct haplotypes among the South American populations. Zardi et al. (2007), sequencing COI, found two genetic lineages (a western and an eastern one) of the brown mussel Perna perna on the South-East coast of South Africa that could be explained by the pattern of ocean currents in that region. Populations of bivalves from other families have also been studied from the Brazilian coast using COI gene sequences. Arruda et al. (2009) found heterogeneity among four populations of Anomalocardia brasiliana, whereby one population presented isolation by distance (Ilha Canela, Bragança) and another isolation due to physical barriers (Camurupim). Population studies carried out by Lazoski et al. (2011) with allozymes and COI sequences, show intraspecific genetic structure in the oysters Crassostrea brasiliana and Crassostrea rhizophorae, which, according to the authors, follows the isolation by distance model. There are only a few genetic studies of mussels of the genus Mytella (Oliveira et al., 2005; Gillis et al., 2009; Alves et al., 2012). The present study is the first based on molecular data (COI and 18SITS1) from a wide range of populations along the Brazilian coast and the objective of this paper is to investigate the genetic diversity, population structure, and demographic history of the native mussel species Mytella charruana. Moreover, our study aims to identify useful DNA markers for genetically characterizing the species M. charruana. As three very distinct groups of COI sequences were found, this led us to investigate the possibility of the presence of cryptic species, an ancient polymorphism and/or doubly uniparental inheritance, all of which have previously been described in Foighil, 2004; Sirne Terranova et al., 2006, the Mytilidae (Lee and O 2007; Alves et al., 2012). 2. Material and methods A total of 243 individual mussels were sampled at 10 localities along the Brazilian coast, from Oiapoque on the northern coast, to Antonina, on the eastern coast. The sample size ranged from 15 to 30 at each locality (Table 1). Whole specimens were conserved in 92% ethanol and frozen at 20 C pending laboratory analyzes. In order to determine which COI haplotypes are patrilinear and which are matrilinear, the sex of each individual was determined in six specimens of Mytella charruana from Bragança and ten from Maceio using hematoxilin-eosin dye and the methods described by Alves et al. (2012). The sex of charru mussels from Antonina (N ¼ 10) was determined by using fresh gonad smears by adding one drop of distilled water to the gonad material, which was examined under the light microscope (400). All mussels were morphologically identified to species and the shells were deposited in the Laborio de Conservaça ~o e Biologia Evolutiva of the Universidade rato (LCBE/UFPA) or in the Museu de Zoologia da UniFederal do Para ~o Paulo (MZSP). versidade de Sa DNA was extracted from adductor muscles using the phenolchloroform protocol of Sambrook and Russell (2001). DNA from gonads was extracted when the muscle sequences (mitochondrial Please cite this article in press as: de Souza, T.O., et al., Population structure and identification of two matrilinear and one patrilinear mitochondrial lineages in the mussel Mytella charruana, Estuarine, Coastal and Shelf Science (2014), http://dx.doi.org/10.1016/j.ecss.2014.11.009 T.O. de Souza et al. / Estuarine, Coastal and Shelf Science xxx (2014) 1e10 3 Table 1 Sampling locations, geographic coordinates, number of specimens in each haplogroup, number of haplotypes, and haplotype and nucleotide diversities for the ten populations of Mytella charruana from the Brazilian coast, based on COI gene sequences. Locations Coordinates Samples (N) Specimens from haplogroup A (NA) Specimens from haplogroup B (NB) Number of distinct haplotypes (NH) Haplotype diversity (h) Nucleotide diversity (p) Oiapoque ~o Joa ~o de Pirabas (1) Sa Bragança (2) ^a (3) Augusto Corre Viseu (4) Northern Coast (1 þ 2þ3 þ 4) Fortim Cabedelo Goiana Maceio Antonina 04 020 0800 N51 120 000 W 00 460 08.8700 S47 100 47.0800 W 00 520 2600 S46 380 5900 W 01 010 12.1400 S46 390 12.6800 W 01 000 38.8200 S46 200 4.8600 W e 02 510 25.3200 S40 050 28.500 W 06 580 4900 S34 490 4900 W 07 350 S34 500 W 09 380 00.2500 S35 460 00.1900 W 25 270 1700 S48 400 43.400 W 25 22 24 26 15 87 29 23 25 30 24 25 22 24 26 15 87 0 0 21 5 1 0 0 0 0 0 0 29 23 4 25 23 2 5 11 11 6 28 4 2 10 6 2 0.153 0.407 0.670 0.674 0.571 0.587 0.502 0.087 0.720 0.703 0.083 0.0003 0.0011 0.0032 0.0023 0.0016 0.0021 0.0024 0.0002 0.0190 0.0199 0.0052 COI) suggested heteroplasmy and from individuals of known sex. Part of the mitochondrial COI gene was amplified using the polymerase chain reaction (PCR) with primers designed by Folmer et al. (1994). COI sequences were compared with Mytella charruana sequences deposited in the GenBank (JQ685156 to JQ685159) by Alves et al. (2012) to identify those belonging to the matrilinear or patrilinear lineages. To identify cryptic species, especially in families of bivalves that are known to have DUI, a nuclear gene must be sequenced in order to confirm if differences observed among the mitochondrial sequences represent evolution of the species under study or simply that of the mitochondrial genome. The nuclear DNA fragment, the 18SrRNA (partial) and the internal transcribed spacer 1 (18S-ITS1), was chosen as the latter is already recognized as a useful species specific marker in Mytilidae (Santaclara et al., 2006; Wood et al., 2007; Alves et al., 2012). A subsample of 29 individuals of both sexes from three COI haplogroups (NA ¼ 14; NB ¼ 6; NC ¼ 9) was sequenced for 18S-ITS1 with primers designed by Pleyte et al. (1992) and the protocols used for the amplification of the COI and 18S-ITS1 regions are described in Alves et al. (2012). Amplification reaction products were sequenced for both strands on an ABI 3500 (Applied Biosystems) using the dideoxiterminal florescent marker (BigDye® Terminator v3.1 Cycle Sequencing Kit) according to the manufacturer's instructions. DNA sequences were edited and aligned using Clustal W (Thompson et al., 1997) in BioEdit 7.1.11 (Hall, 1999). The haplotype network was obtained using COI sequences of mussels sampled in the present study, in addition to sequences of Mytella charruana deposited by Gillis et al. (2009) in GenBank (EU91742 to EU917180) from Colombia, Ecuador and the USA. Haploviewer (Salzburger et al., 2011) was used to construct the COI haplotype network in order to to establish the relationship between different haplotypes considering only variable sites. M and F lineages in COI sequences were identified by direct comparison with COI sequences in GenBank (JQ685156, JQ685159) published by Alves et al. (2012), which showed p-distances ranging from 20.5% to 20.8%. In our study, COI sequences were found to belong to both the mtDNA F and mtDNA M lineages and those belonging to the latter were removed from population analyzes since these evolve independently (Hoeh et al., 1996; Breton et al., 2007). Nucleotide frequencies, termination codons and changes in amino-acids were verified with Mega 5.0 (Tamura et al., 2011). Identification of haplotypes, grouping of individuals by population, and the generation of the file to be run in Arlequin 3.5.1.2 (Excoffier and Lischer, 2010) were carried out using DNASp 4.1 (Rozas et al., 2003). Arlequin 3.5.1.2 (Excoffier and Lischer, 2010) was further used for the following analyzes. The indexes of haplotype and nucleotide diversities were calculated, and the former is defined as the probability that two randomly chosen haplotypes are different in the sample and the latter as the probability that two randomly chosen homologous nucleotide sites are different (Nei, 1987). Tajima's (D, Tajima, 1989) and Fu's (FS, Fu, 1997) tests were used to check if the population was in a state of selective neutrality. Negative and significant values of D (Tajima's test) are suggestive of purifying selection, recent increase in population size, and population subdivision (Tajima, 1993). Negative and significant values of FS (Fu's test) indicate recent population expansion or hitchhiking (Fu, 1997). The hypothesis of mussel population expansion in the present study was investigated by analysis of the distribution of pairwise differences in the population (mismatch distribution). A unimodal mismatch distribution is evidence that the population is going through a rapid and sudden expansion (Rogers and Harpending, 1992). The significance of the differences between these values was calculated from the sum of squared deviations (SSD, Excoffier and Schneider, 1999) with 10,000 permutations. The estimates of the time of expansion were made using the equation t ¼ 2ut (Rogers and Harpending, 1992) and calibrated with a divergence rate of 0.52% per nucleotide per million years estimated for COI by Luttikhuizen et al. (2003) who used bivalve fossil record dates. The generation time was estimated at 1 year (Pereira et al., 2003). Genetic structure among populations was tested by pairwise FST and significance was checked using 10,000 permutations. The partitioning of genetic variability was tested using analysis of molecular variance (AMOVA, Excoffier and Lischer, 2010) with 10,000 permutations. Both previous analyzes were developed in Arlequin 3.5.1.2 (Excoffier and Lischer, 2010). AMOVA was carried out considering all populations together; and, due to the clear separation, by at least 30 mutations, of the matrilinear lineage in two groups (haplogroups A and B) from Brazilian populations, by pooling the populations in their haplogroups. Associations among FST values and coastal geographic distances among Mytella charruana populations were tested considering all populations together, and, separated by haplogroups. For this purpose, the Mantel test (10,000 permutations) was carried out using IBDWS online (Jensen et al., 2005). Geographic distances between populations were estimated using Google Earth by measuring the distance along the coastline. The evolutionary model and the parameters used to obtain the maximum likelihood (ML) tree were selected by JModeltest (Guindon and Gascuel, 2003; Posada, 2008). The significance of the ML tree arrangements were tested by bootstrapping with 1000 pseudo-replicates using PhyML 3.0 (Guindon et al., 2010). Bootstrap values equal or superior to 95% were considered statistically significant (Li, 1997). COI sequences from other Mytilidae were added to the phylogenetic analysis (Genbank numbers: JQ685160, JQ685162, GU570464, AY497292, JX486124). Please cite this article in press as: de Souza, T.O., et al., Population structure and identification of two matrilinear and one patrilinear mitochondrial lineages in the mussel Mytella charruana, Estuarine, Coastal and Shelf Science (2014), http://dx.doi.org/10.1016/j.ecss.2014.11.009 4 T.O. de Souza et al. / Estuarine, Coastal and Shelf Science xxx (2014) 1e10 A phylogenetic tree based on Bayesian Inference (BI), and using COI sequences, was created in BEAST 2 (Bouckaert et al., 2014) to estimate the time to the most recent common ancestor (tMRCA) among mtDNA lineages of Mytella charruana and other Mytilidae. The input file for BEAST 2 was created using the application BEAUti. The phylogenetic tree used the strict clock model and the prior was set to the default option of the Yule process (Drummond et al., 2006). Fossil dating (Barsotti and Meluzzi, 1968; Coan et al., 2000) of the separation of Mytilus galloprovincialis and Mytilus edulis (2 Mya); and Mytilus californianus and M. edulis - M. galloprovincialis (30 Mya) were used as references to calculate the tMRCA. 3. Results In subsamples of Mytella charruana (N ¼ 26) there were four females and two males in Bragança; eight females, one male and ; and one male, seven females one of indeterminate sex in Maceio and two individuals of indeterminate sex in Antonina. All specimens sequenced in this study were morphologically identified as M. charruana, and the shells from all localities were deposited as lots 109537 to 109545 and 109546 to 109551 in MZUSP and in LCBE/UFPA. Sequences of the nuclear fragment 18S-ITS1 agree with this identification when compared with the sequence JQ734971 from GenBank (Alves et al., 2012), with reference to lot number 99629 at MZUSP. The 18S-ITS1 showed four very similar sequences with only one mutation and an indel of 1e2 nucleotides and were not related to any COI haplogroups. We unambiguously sequenced 510 bp of the COI gene (mtDNA F lineage) from a total of 243 Mytella charruana and the COI sequences could clearly be divided into three very distinct haplogroups: A, B and C (Fig. 1). Comparison with sequences deposited in GenBank (JQ685158, JQ685159) by Alves et al. (2012), made possible the unambiguous identification of a patrilinear lineage (haplogroup C) in the COI sequences of specimens collected in Oiapoque (N ¼ 12), Bragança (N ¼ 4), Viseu (N ¼ 4), Cabedelo (N ¼ 2) and Antonina (N ¼ 1). The (N ¼ 2), Goiana (N ¼ 1), Maceio comparison of COI sequences from mtDNA M (patrilinear) and mtDNA F (matrilinear) lineages presented between 103 and 107 mutated sites (Fig. 1) and p-distances of 0.2081 and 0.2097, respectively. The same patrilinear lineage (haplogroup C) was found in males with both matrilinear lineages (haplogroup A and B). Haplogroup C had eight distinct haplotypes (H47 to H54) with two to seven nucleotides and one to two amino acid mutations between them. COI sequences were obtained from a total of 243 Mytella charruana, varying from 15 to 30 individuals at each of the 10 localities (Table 1). Among all populations, 46 haplotypes were identified from the mtDNA F lineage (GenBank accession numbers KP013759 to KP013804), which were classified, according to the network division, into two distinct haplogroups (Fig. 1). As seen in Fig. 2, haplogroup A was predominant in mangroves from northern Brazil, from Oiapoque to Viseu, whereas haplogroup B was predominant from Fortim (northeast) to Antonina (east), with the exception of Goiana (northeast) where sequences from haplogroup A predominated (N ¼ 21) over those from B (N ¼ 4). Haplogroup A was separated from haplogroup B by at least 30 mutations (p-distance: 0.0624). In haplogroup A, 38 haplotypes were found, of which H1 was the most frequent (N ¼ 79), followed by H30 (N ¼ 19). In haplogroup B, a total of 8 haplotypes was found, of which H39 (N ¼ 60) was the most common, followed by H44 (N ¼ 23) and H41 (N ¼ 12). The remaining haplotypes of both haplogroups were found in sequences from 1 to 3 individuals. Within haplogroups, the number of mutations varied from 1 to 7 for group A and from 1 to 4 for B (Fig. 1). Gillis et al. (2009) COI sequences of M. charruana from GenBank (EU917142 to EU917180) were related to haplogroup B (Figs. 1 and 2), which predominated in eastern Brazilian populations, but, however, did not share haplotypes with any of the Brazilian populations. After inclusion of the haplotypes from Gillis et al. (2009) study, haplogroups A and B were separated by 26 mutations. Therefore, based on the network arrangements, the M. charruana COI sequences were divided into two matrilinear haplogroups (A and B) and one patrilinear (C). In haplogroup A, the haplotype distribution from northern Brazil and from Goiana exhibit a star shaped network suggestive of recent demographic expansion (Grant and Bowen, 1998). Fig. 1. The haplotype network divided the COI sequences (510 bp) from 10 populations of Mytella charruana from the Brazilian coast into two matrilinear and one patrilinear groups, and showed that the matrilinear group B is also present in sequences from Colombia, Ecuador and the United States of America (USA) obtained from Gillis et al. (2009). H1 to H46: Brazilian matrilinear haplotypes. Letters: Haplotypes from Colombia, Ecuador and USA. Please cite this article in press as: de Souza, T.O., et al., Population structure and identification of two matrilinear and one patrilinear mitochondrial lineages in the mussel Mytella charruana, Estuarine, Coastal and Shelf Science (2014), http://dx.doi.org/10.1016/j.ecss.2014.11.009 T.O. de Souza et al. / Estuarine, Coastal and Shelf Science xxx (2014) 1e10 5 Fig. 2. Sampling locations of Mytella charruana and proportions of haplogroups A and B at each location. In the Antonina population (east coast), 23 individuals had haplotype H44, found only at this locality and belonging to haplogroup B, and one specimen had haplotype H30, from haplogroup A. Individuals from the east coast of Brazil from haplogroup A , N ¼ 5; Antonina, N ¼ 1) do not share the (Goiana, N ¼ 21; Maceio same haplotypes with individuals from the north coast, there being at least two mutations between them. The global analysis showed intrapopulation haplotype and nucleotide diversities ranging from 0.083 (Antonina) to 0.720 ), respectively (Goiana), and 0.0002 (Cabedelo) to 0.0199 (Maceio and Antonina, (Table 1). Only three populations, Goiana, Maceio showed haplotypes from both haplogroups (A and B). A second analysis, considering only haplotypes belonging to the predominant haplogroup was carried out for these populations and, in this case, the haplotype diversities ranged from 0.000 (Antonina) to 0.629 (Goiana) and the nucleotide diversities from 0.0000 (Anto ). The nucleotide diversities were low nina) to 0.0030 (Maceio among haplotypes of the same haplogroup but high in comparison to those of other haplogroups. Genetic structure (FST) was found among the majority of pairwise populations using all haplotypes present in the populations and using only haplotypes of the predominant haplogroup in each population (Table 2). AMOVA among all populations and considering all haplotypes corroborated these results showing that 82.07% of the variation was among populations (FST ¼ 0.820; P < 0.001). ~o Joa ~o However, populations from the northern coast, between Sa de Pirabas and Viseu, were homogenous (FST ¼ 0.005; P ¼ 0.227), but when Oiapoque is added to this group, AMOVA indicates heterogeneity (FST ¼ 0.013; P ¼ 0.050). AMOVA between haplogroups A and B showed heterogeneity, responsible for 85.84% of the variation and also among the populations within both groups F (FCT ¼ 0.858; P ¼ 0.004). When genetic (FST values) and geographic distances of all populations of Mytella charruana were compared, Mantel test results Table 2 Pairwise FST value comparisons among populations of Brazilian Mytella charruana. Lower diagonal: the analysis was carried with all individuals from each population. Upper diagonal: only individuals from the predominant haplogroup were used in each population. Locations Oiapoque ~o de Pirabas S~ ao Joa Bragança ^a Augusto Corre Viseu Goiana Fortim Cabedelo Maceio Antonina Oiapoque ~o Joa ~o de Pirabas Sa Bragança ^a Augusto Corre Viseu Goiana Fortim Cabedelo Maceio Antonina e 0.02560 0.03732* 0.02975 0.03017 0.29046* 0.97573* 0.99595* 0.77984* 0.95212* 0.02560 e 0.01445 0.00453 0.01109 0.26573* 0.96910* 0.98979* 0.76357* 0.94305* 0.03732* 0.01445 e 0.00227 0.01408 0.24813* 0.95454* 0.97193* 0.75755* 0.92793* 0.02975 0.00453 0.00227 e 0.00720 0.27061* 0.95997* 0.97770* 0.76626* 0.93478* 0.03017 0.01109 0.01408 0.00720 e 0.22305* 0.96405* 0.98787* 0.73323* 0.93282* 0.81267* 0.74656* 0.59924* 0.66175* 0.70890* e 0.81009* 0.81442* 0.57721* 0.76632* 0.97573* 0.96910* 0.95454* 0.95997* 0.96405* 0.96717* e 0.13016* 0.12852* 0.44002* 0.99595* 0.98979* 0.97193* 0.97770* 0.98787* 0.98626* 0.13016* e 0.16801* 0.57617* 0.97155* 0.96401* 0.94834* 0.95445* 0.95752* 0.96194* 0.05349 0.31709* e 0.19592* 0.99734* 0.99129* 0.97335* 0.97907* 0.98965* 0.98769* 0.69460* 0.97872* 0.66536* e *P < 0.05. Please cite this article in press as: de Souza, T.O., et al., Population structure and identification of two matrilinear and one patrilinear mitochondrial lineages in the mussel Mytella charruana, Estuarine, Coastal and Shelf Science (2014), http://dx.doi.org/10.1016/j.ecss.2014.11.009 6 T.O. de Souza et al. / Estuarine, Coastal and Shelf Science xxx (2014) 1e10 Table 3 Populations of Mytella charruana from the Brazilian coast classified by haplogroup with details of sample size, number of haplotypes, and results of Tajima's (D) and Fu's (Fs) tests, sum of squared deviations (SSD) and tau (t). t Locations N Haplotypes (N) D (P) FS (P) SSD (P) Oiapoque ~o Jo~ Sa ao de Pirabas (1) Bragança (2) ^a (3) Augusto Corre Viseu (4) Homogenous Northern Coast populations (1þ2þ3þ4) Fortim Cabedelo Goiana Maceio Antonina Only haplotypes from Haplogroup A Goiana Only haplotypes from Haplogroup B Maceio Antonina 25 22 24 26 15 87 29 23 25 30 24 2 5 11 11 6 28 4 2 10 6 2 0.69827 (0.22780) 1.80901 (0.01510) 2.00002 (0.00840) 2.08909 (0.00550) 1.98306 (0.00820) 2.55797 (0.00000) 0.11134 (0.49060) 1.16097 (0.14650) 0.24363 (0.45270) 0.65598 (0.79310) 2.60844 (0.00010) 0.28345 (0.17260) 2.66224 (0.00740) 6.46281 (0.00010) 8.04625 (0.00000) 3.53713 (0.00080) 29.68505 (0.00000) 0.88475 (0.71000) 0.99304 (0.07140) 2.45687 (0.85030) 9.31976 (0.99400) 7.17462 (0.99440) 0.02833 0.00066 0.01395 0.00036 0.00035 0.00045 0.35448 0.00003 0.05624 0.10793 0.01026 (0.10630) (0.81150) (0.76950) (0.95910) (0.82490) (0.95830) (0.95510) (0.81460) (0.41090) (0.11640) (0.84350) 2.965 0.500 2.961 1.447 0.848 1.916 0 3.000 1.078 31.586 3.000 21 9 1.84558 (0.01850) 7.53331 (0.00000) 0.00707 (0.42600) 0.955 25 23 5 1 1.20908 (0.89000) 0.00000 (1.00000) 0.28718 (0.59260) 0.00000 (N.A) 0.10957 (0.14830) 0.00000 (0.00000) 3.629 0 were significant (r ¼ 0.533, P ¼ 0.005). When separated by haplogroup, results were significant only for haplogroup A (r ¼ 0.905, P ¼ 0.002), in contrast to haplogroup B (r ¼ 0.800, P ¼ 0.078). Tajima's (D) and Fu's (FS) values (Table 3) revealed deviation from neutrality with significant negative values in four populations ~o Joa ~o de Pirabas, Bragança, Augusto Corre ^a from northern Brazil (Sa and Viseu). As the latter four populations were homogenous, these were grouped and both neutrality tests were recalculated, again resulting in significant negative values. Antonina, located on the east coast, deviated only for Tajima's test, probably due to a bottleneck. The remaining populations did not present deviation from selective neutrality. In three populations, individuals from both haplogroups were found (Table 3). In Goiana, specimens from and Antohaplogroup A predominated (84%) whereas in Maceio nina, haplogroup B was the most frequent (83.3% and 95.8%, respectively). A large difference in the frequency of one haplogroup with respect to another in a population suggests recent mixture of stocks, therefore Fu's and Tajima's tests were also carried out without the haplotypes of the minority haplogroup for Goiana, and Antonina. The latter analysis showed deviation of Maceio neutrality and signs of demographic expansion only for Goiana (Table 3). The northern coast group, comprising the four homogenous populations, and Goiana (only haplogroup A) both showed unimodal pairwise mismatch distributions, with curves indicating recent expansion (Fig. 3a and b, respectively). The sum of squared deviations (SSD) analysis (Table 3) supported the hypothesis of recent demographic expansion in the northern coast group and in the Goiana population. Considering a mutation rate of 0.52% (Luttikhuizen et al., 2003), the northern coast group showed signs of demographic expansion, which began around 360,000 years ago, and Goiana, in the northeast, presented demographic expansion dating from 180,000 years ago. The Bayesian coalescent phylogenetic tree (Fig. 4) shows that the coalescent time calculated for Mytella charruana was 12.5 Mya; for haplogroup A 3.4 Mya and for haplogroup B 4.0 Mya; for northern populations 2.8 Mya, and, for the Goiana population 1.9 Mya. The patrilinear lineage of M. charruana coalesced prior to the origin of Perna perna (51.2 Mya). The evolutionary model selected by JModeltest (Guindon and Gascuel, 2003; Posada, 2008) for the Mytella charruana haplotypes was the K81 (Kimura, 1981) method and the proportion of invariable sites was 0.7040. A Maximum Likelihood (ML) analysis was ran and resulted in a phylogenetic tree with topology similar to thatof the BI tree (Fig. 4), thus we included the bootstrap values of ML in the BI tree. The arrangements of ML and BI trees supported the results shown in the haplotype network (Fig. 1), dividing the COI sequences into three haplogroups supported by bootstrap values ranging from 92% to 100%. The relationships among matrilinear M. charruana, M. guyanensis and Perna perna were not supported by strong bootstrap values, however the basal position of the patrilinear lineage is clear (bootstrap: 100%). 4. Discussion COI sequences from Mytella charruana revealed two matrilinear lineages and one patrilinear lineage. Mitochondrial sequence heteroplasmy was found in populations of both haplogroups of Mytella Fig. 3. Observed distribution of pairwise differences, in comparison to the expected population growth model, based on COI gene sequences from Brazilian Mytella charruana. a) four homogenous populations from the northern coast; b) population from Goiana (northeastern coast), including only haplotypes belonging to haplogroup A. Please cite this article in press as: de Souza, T.O., et al., Population structure and identification of two matrilinear and one patrilinear mitochondrial lineages in the mussel Mytella charruana, Estuarine, Coastal and Shelf Science (2014), http://dx.doi.org/10.1016/j.ecss.2014.11.009 T.O. de Souza et al. / Estuarine, Coastal and Shelf Science xxx (2014) 1e10 7 Fig. 4. Bayesian coalescent phylogenetic tree with the estimate of the time to the most recent common ancestor assuming a strict molecular clock, based on COI sequences for haplogroups A, B and C of M. charruana and sequences of other Mytilidae with dates of the separation in million of years ago and confidence interval in parentheses. A previous Maximum Likelihood analysis resulted in a phylogenetic tree with similar topology and bootstrap values (1000 pseudo-replicates) are shown preceding an asterisk. charruana and comparisons with Alves et al. (2012) sequences indicate the presence of DUI. As the patrilinear lineage of the mtDNA evolves faster than the matrilinear (Passamonti and Ghiselli, 2009; Zouros, 2013), only the latter is used in population analyzes (Hurlwood et al., 2005; Arruda et al., 2009; Gillis et al., 2009; Lazoski et al., 2011). found in the 18S-ITS1 sequences may be related to concerted evolution of repetitive genes with consequent homogenization of this fragment (Elder-Jr and Turner, 1995). The absence of a second patrilinear mitocondrial lineage may be due a lack of specificity of the pair of primers used, perhaps because of a mutation peculiar to this lineage. 4.1. Two matrilinear haplogroups: one species or two cryptic species? 4.2. Geographic distribution of the haplogroups The large differences among COI mitochondrial sequences from the matrilinear lineages from Brazil resulted in haplotypes being classified into two distinct haplogroups. Most populations had moderate haplotype diversity and low nucleotide diversity, except , in which haplotypes from the two distinct Goiana and Maceio haplogroups A and B are established, and in this case both haplotype and nucleotide diversities are high (Table 1). Although there was no evidence of morphological differences among the Mytella charruana sampled from the 10 localities along the Brazilian coast, the presence of two mitochondrial haplogroups could suggest the existence of two cryptic species. However, nuclear sequences of the 18S-ITS1 fragment, considered a useful marker for mytilid species (Santaclara et al., 2006; Wood et al., 2007), did not reveal significant differences among the 18S-ITS1 sequences of specimens of both matrilinear (A and B) and patrilinear (C) COI haplogroups, thus supporting a single species hypothesis for M. charruana. Moreover, the patrilinear sequences revealed only one haplogroup, C, present in all populations also indicating that northern and northeasterneastern populations belong to the same species. Even if a speciation process has begun in the charru mussel, either by geographic isolation or ecological adaptation, the present data suggest that individuals of different COI haplogroups are still able to mix, since they share alleles from the 18S-ITS1 fragment. The low variability, even after an initial diversification process, The differences between the two matrilinear haplogroups of Mytella charruana from the Brazilian coast also appear to be related to geographic distribution, with the unique presence of one group on the northern coast (haplogroup A) and the other predominant along the northeastern and eastern coasts (haplogroup B). In populations of Perna perna from eastern and western South Africa, a lesser degree of divergence, involving approximately 12 mutations, was detected using COI sequences (400 bp) and a central zone, where overlap occurred, was evident (Zardi et al., 2007). According to the authors, maritime currents or local selective forces would explain these differences. Similar results were found in comparisons of sequences of specimens of P. perna from South Africa with those of Morocco and Mauritania, from North Africa (Wood et al., 2007). Another species with two COI haplotype lineages is the Pacific oyster or Japanese oyster Crassostrea gigas, which, when brought to Europe probably by accident, became known as Crassostrea angasi (Hurwood et al., 2005). However, despite the differences, the number of mutations in COI sequences (547 bp) between both stocks, also around 12, does not justify their classification as two distinct species (Reece et al., 2008). The samples sequenced from Colombia by Gillis et al. (2009) are related to haplogroup B, found in northeastern and eastern Brazil, and not to populations from northern Brazil that are, in fact, geographically closer, suggesting overlap of the distribution of the haplogroups. However, in the present study, five locations were Please cite this article in press as: de Souza, T.O., et al., Population structure and identification of two matrilinear and one patrilinear mitochondrial lineages in the mussel Mytella charruana, Estuarine, Coastal and Shelf Science (2014), http://dx.doi.org/10.1016/j.ecss.2014.11.009 8 T.O. de Souza et al. / Estuarine, Coastal and Shelf Science xxx (2014) 1e10 sampled around the mouth of the Amazon river, between Oiapoque and Viseu, and no evidence of haplotypes from haplogroup B was found in this region. Comparisons among sequences from the maternal lineage with those of Gillis et al. (2009), showed similarities between the Brazilian populations with haplogroup B, predominant along the northeastern and eastern coasts, and four invasive populations in the United States and two native ones, from Cartagena (Colombia) and Guayaquil (Ecuador). However, none of the Brazilian COI sequences were identical to those of Mytella charruana in Gillis et al. (2009) suggesting that Brazilian stocks were not involved in the invasions studied by the latter authors. The specimens collected from Goiana (northeastern coast) were from haplogroup A. However, this population was significantly different from the remaining populations in this group, located on the northern coast, showing that this is a distinct population, probably due to a founder effect and isolation by distance. The direction of the maritime currents are from the northeast to the North, via the North Brazil Current (Cirano et al., 2006), however, only one individual from the northern coast (Bragança), presented a haplotype (H16) derived from the Goiana stock (Fig. 1). This finding suggests that the charru mussel from haplogroup B (predominant in the Northeast and East) may be unable to survive in the mangroves of the North, which receive high freshwater discharges during the rainy season. Salinity, in 2003, ranged from 10.9 to 40 in the Bragança region, for example (Santos-Filho et al., 2008). On the other hand, the population of the mussel Brachidontes pharaonis from Salina di Marsala (Mediterranean Sea, Italy) had private haplotypes in high frequencies that may be selectively advantageous in waters with high temperature and salinity (SirnaTerranova et al., 2006). 4.3. Demographic history All northern populations from haplogroup A, except those from Oiapoque, were homogenous and showed evidence of population expansion dating from around 360,000 years ago. Moreover, the Goiana Mytella charruana stock, which also belongs to haplogroup A, showed significant population growth around 180,000 years ago. Over the past 700,000 years, Earth has been subject to widespread and long-lasting glaciations interrupted by shorter, warmer interglacial periods that have lead to successive changes in sea level, altering the shape of the coastline and the distribution of coastal species (Hewitt, 1996). Consequently, many populations may have suffered bottlenecks, been permanently isolated or accumulated genetic differences by drift or selection for local adaptations. Currently, rising CO2 concentrations are changing the temperature and the acidity of the ocean and, along with changes in salinity with variable freshwater inflow, relative sea level rise is redesigning the coast line in many parts of the world (Roessig et al., 2004). The populations from the northeastern and eastern coasts, where haplogroup B predominated, are in selective neutrality, except for Antonina, where there was evidence of a bottleneck or founder effect. In this locality, the exact sampling location, in the mangrove, was relatively protected from the influence of oceanic currents that may limit natural dispersion in the region. Cho et al. (2007) found similar results with six populations of Scapharca broghtonii from Korea that presented genetic differences among which a single population (Jinhae) was differentiated in relation to the others since it had limited dispersal with the presence of a narrow avenue for exchange with marine currents. On the other hand, the sampling location in Antonina was close to a busy port and the presence of a single haplotype from haplogroup B (N ¼ 23) and another from haplogroup A (N ¼ 1) may be explained by their recent introduction through shipping. The transport of incrusting bivalves on hulls and in ballast water is well known and has been documented by Farrapeira et al. (2011) for the Brazilian coast. With the current data, it was not possible to establish with certainty the cause of the separation of Mytella charruana into two mitochondrial haplogroups. The coalescence of Atlantic charru mussels goes back a long time, around 12.5 Mya (CI: 8.3 to 17.3 Mya); and, 3.4 Mya (CI: 2 to 5 Mya) and 4 Mya (CI: 2.7 to 5.7 Mya) for haplogroups A and B, respectively. This was a period of important changes when excessive global cooling severely reduced the sea level and when, around 10.5 Mya, the direction of the Amazon river was inverted (Figueiredo et al., 2009), which may have drastically affected the ecosystem around its new mouth. Although the changes in amino acids produced by the nucleotide mutations in the COI gene were sporadic and did not represent important differences between the haplogroups, the A lineage mainly found around the mouth of the Amazon river, may represent adaptations to low salinity, high turbidity and other factors characteristic of this coastal area. 4.4. Doubly-uniparental inheritance Considering the existence of DUI in this species (Alves et al., 2012), the hypothesis of role-reversals (Hoeh et al., 1997) may arise. Could the presence of two matrilinear lineages be explained by an unusual feminization of a mtDNA M? Could the absence of a second patrilinear lineage be due to a recent masculinization of the mtDNA F? Recent studies based on sequences of the control region (CR) of Mytilus species showed large differences among matrilinear and patrilinear mitochondrial sequences, the results indicated that role-reversal is a rare event involving recombination of both genomes (Zouros, 2013). Moreover, feminization of the male genome has never been described in bivalves, the matrilinear mtDNA genome is under stronger selection than the patrilinear one, which allows more frequent amino acid mutations that may eventually be fixed in the latter lineage. It provides an explanation as to why recent mtDNA masculinized genomes (F to M) have a higher chance of success than feminized ones (M to F) (Hoeh et al., 1997; Zouros, 2013). Comparison of our data with those of Alves et al. (2012), shows that, besides the high nucleotide mutation rates observed for the COI gene of patrilinear origin, they also found a high rate of change in amino acids, which was not found in lineages of both matrilinear haplogroups in our study. Therefore, the data do not support the occurrence of role-reversal from mtDNA. 4.5. Independent mitochondrial evolution or interrupted speciation? With the peculiar pattern of mitochondrial inheritance, where recombination does not occur and the mitochondria are sent directly from the mother to her offspring, lineages may evolve independently until they disappear due to drift or natural selection (Avise, 2000). However, if the population is numerous and the mutations result in adaptive effects that do not lead to disadvantages, the population may retain more than one mitochondrial lineage evolving independently for many generations. This last hypothesis is probably the cause of the observed divergence. 4.6. Conclusions and advice for management The present study, based on a substantial number of populations and specimens, shows that mtDNA genes can be important tools in evolutionary studies, but also that they should be used with caution due to their particular pattern of matrilinear and non recombinant inheritance, in addition to doubly-uniparental inheritance in some species. The extensive study allows us to suggest with confidence, Please cite this article in press as: de Souza, T.O., et al., Population structure and identification of two matrilinear and one patrilinear mitochondrial lineages in the mussel Mytella charruana, Estuarine, Coastal and Shelf Science (2014), http://dx.doi.org/10.1016/j.ecss.2014.11.009 T.O. de Souza et al. / Estuarine, Coastal and Shelf Science xxx (2014) 1e10 the origin in time of the three distinct lineages (haplogroups) of COI sequences. Two were matrilinear, separated from each other probably around 3.4 and 4 Mya, and one patrilinear, which split from the matrilinear lineage around 51.2 Mya, before the separation of the genera Perna and Mytella. The correct identification of COI haplogroups and their haplotypes in charru mussels now allow these sequences to be used with confidence for taxonomic purposes and in management for aquaculture and conservation. Only haplogroup A is found around the Amazon river discharge, suggesting that there is some environmental factor in that area selecting against individuals from haplogroup B. Differences in salinity and temperature may have selected for distinct lineages of mussels (Sirna-Terranova et al., 2006) and more detailed data on temperature, pH, salinity and local currents could help explain the genetic structuring observed among populations of Brazilian Mytella charruana. Global warming is changing the coast line and changing the freshwater inflow into estuaries that may cause severe bottlenecks effects, especially in estuarine species (Roessig et al., 2004). Knowledge of the genetic diversity of the species and their tolerance to environmental changes is necessary and urgent to plan strategies to minimize local impacts of global warming, especially when a considerable number of the involved species are used as a food source, such as fishes and mussels. Acknowledgments We thank the Conselho Nacional de Desenvolvimento Científico gico (CNPq) for funding this research via a grant (Edital e Tecnolo Universal 2009, Grant No. 472558/2009-9) and a scholarship (552344/2010-9) awarded to T. Souza. Sampling was carried out under License No. 21823-1 from the Instituto Chico Mendes de ~o da Biodiversidade (ICMBIO). We would like to thank Conservaça Mauro Melo and Jaqueline Oliveira for collecting some of the samples used in this paper. Appendix A. Supplementary data Supplementary data related to this article can be found at http:// dx.doi.org/10.1016/j.ecss.2014.11.009. References Alves, F.A.S., Beasley, C.R., Hoeh, W.R., Rocha, R.M., Simone, L.R.L., Tagliaro, C.H., 2012. Detection of mitochondrial DNA heteroplasmy suggests a doubly unipa^nc. rental inheritance pattern in the mussel Mytella charruana. Rev. Bras. Biocie 10, 176e185. Arruda, C.C.B., Beasley, C.R., Vallinoto, M., Marques-Silva, N.S., Tagliaro, C.H., 2009. Significant genetic differentiation among populations of Anomalocardia brasiliana (Gmelin, 1791), A bivalve with planktonic larval dispersion. Gen. Mol. Biol. 32, 423e430. .org/10.1590/S1415-47572009000200033. Avise, J.C., 2000. Phylogeography, the History and Formation of Species. Harvard University Press, Cambridge, MA. Barsotti, G., Meluzzi, C., 1968. Osservazioni su Mytilus edulis L. e Mytilus galloprovinicialis Lamarck. Conchiglie Milan. 4, 50e58. Bouckaert, R., Heled, J., Kühnert, D., Vaughan, T.G., Wu, C.-H., Xie, D., Suchard, M.A., Rambaut, A., Drummond, A.J., 2014. BEAST2: a software platform for Bayesian evolutionary analysis. PLOS Comput. Biol. 10 (4) http://dx.doi.org/10.1371/ journal.pcbi.1003537. , H.D., Stewart, D.T., Hoeh, W.R., Blier, P.U., 2007. The unusual Breton, S., Beaupre system of doubly uniparental inheritance of mtDNA, isn't one enough? TIG 23, 465e474. http://dx.doi.org/10.1016/j.tig.2007.05.011. Brodsky, S., Walters, L.J., Hoffman, E., Schneider, K., 2009. Thermal tolerances of the invasive mussel Mytella charruana. In: Proceedings of the Society for Integrative and Comparative Biology Annual Meeting, Boston MA. Available at: https//ncur. weber.edu/ncur/archive/Display_NCUR.aspx?id¼20651 (accessed August 2012). Cardenas, E.B., Aranda, D.A., 2000. A review of reproductive patterns of bivalve mollusks from. Mex. Bull. Mar. Sci. 66, 13e27. Cho, E.S., Jung, C.G., Sohn, S.G., Kim, C.W., Han, S.J., 2007. Population genetic structure of the ark shell Scapharca broughtonii Schrenck from Korea, China, and Russia based on COI gene sequences. Mar. Biotech. 9, 203e216. http:// dx.doi.org/10.1007/s10126.006.6057.x. 9 , N.F.R., 2006. A circulaça ~o ocea ^nica de Cirano, M., Mata, M.M., Campos, E.J.D., Deiro ^ntico Sul com base no modelo de circulaça ~o larga-escala na regi~ ao oeste do Atla global OCCAM. Rev. Bras. Geof. 24 (2), 209e230. http://dx.doi.org/10.1590/ S0102-261X2006000200005. Coan, E.V., Scott, P.V., Bernard, F.R., 2000. Bivalve Seashells of Western North America, Marine Bivalve Mollusks from Arctic Alaska to Baja California. Santa Barbara Museum of Natural History, Santa Barbara, CA. Collin, R., 2001. The effects of mode of development on phylogeography and population structure of North Atlantic Crepidula (Gastropoda, Calyptraeidae). Mol. Ecol. 10, 2249e2262. http://dx.doi.org/10.1046/j.1365-294X.2001.01372.x. Drummond, A., Ho, S.Y.W., Phillips, M.J., Rambaut, A., 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4 http://dx.doi.org/10.1371/ j.pbio.0040088, 699e670. Elder-Jr, J.F., Turner, B.J., 1995. Concerted evolution of repetitive DNA-sequences in eukaryotes. Q. Rev. Biol. 70, 297e320. Excoffier, L., Lischer, H.E.L., 2010. Arlequin suite ver 3.5, a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564e567. http://dx.doi.org/10.1111/j.1755-0998.2010.02847.x. Excoffier, L., Schneider, S., 1999. Why hunter-gatherer populations do not show signs of Pleistocene demographic expansions. Proc. Nat. Acad. Sci. U. S. A. 96, 10597e10602. http://dx.doi.org/10.1073/pnas.96.19.10597. rio, D.O., Amaral, F.D., 2011. Vessel biofouling as an inadFarrapeira, C.M.R., Teno vertent vector of benthic invertebrates occurring in Brazil. Mar. Poll. Bull. 62, 832e839. http://dx.doi.org/10.1016/j.marpolbul.2010.12.014. Figueiredo, J., Hoorn, C., Van Der Ven, P., Soares, E., 2009. Late Miocene onset of the Amazon River and the Amazon deep-sea fan, Evidence from the Foz do Amazonas Basin. Geology 37, 619e622. http://dx.doi.org/10.1130/G25567A.1. Folmer, O., Black, M., Hoeh, W., Lutz, R., Vrijenhoek, R., 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotech. 3, 294e299. Fu, Y.X., 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915e925. Gillis, N.K., Walters, L.J., Fernandes, F.C., Hoffman, E.A., 2009. Higher genetic diversity in introduced than in native populations of the mussel Mytella charruana, evidence of population admixture at introduction sites. Divers. Distrib. 15, 784e795. http://dx.doi.org/10.1111/j.1472-4642.2009.00591.x. Grant, W.S., Bowen, B.W., 1998. Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. J. Hered. 89, 415e426. Guindon, S., Gascuel, O., 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696e704. http:// dx.doi.org/10.1080/106335150390235520. Guindon, S., Dufayard, J.F., Lefort, V., Anisimova, M., Hordijk, W., Gascuel, O., 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59 (3), 307e321. Hall, T.A., 1999. BioEdit, a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95e98. Hewitt, G.M., 1996. Some genetics consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 58, 247e276. http://dx.doi.org/ 10.1111/j.1095-8312.1996.tb01434.x. Hoeh, W.R., Stewart, G.W., Sutherland, B.W., Zouros, E., 1996. Multiple origins of gender-associated mitochondrial DNA lineages in bivalves (Mollusca, Bivalvia). Evolution 50, 2276e2286. Hoeh, W.R., Stewart, D.T., Saavedra, C., Sutherland, B.W., Zouros, E., 1997. Phylogenetic evidence for role-reversals of gender-associated mitochondrial DNA in Mytilus (Bivalvia, Mytilidae). Mol. Biol. Evol. 14 (9), 959e967. http://dx.doi.org/ 10.1093/oxfordjournals.molbev.a025839. Hoeh, W.R., Stewart, D.T., Guttman, S.I., 2002. High fidelity of mitochondrial genome transmission under the doubly uniparental mode of inheritance in freshwater mussels (Bivalvia, Unionoidea). Evolution 56 (11), 2252e2261. http://dx.doi.org/ 10.1111/j.0014-3820.2002.tb00149.x. Hoskin, M.G., 1997. Effects of contrasting modes of larval development on the genetic structures of populations of three species of prosobranch gastropods. Mar. Biol. 127, 647e656. http://dx.doi.org/10.1007/s002270050055. Hurwood, D.A., Heasman, M.P., Mather, P.B., 2005. Gene flow, colonisation and demographic history of the flat oyster Ostrea angasi. Mar. Freshw. Res. 56 (8), 1099e1106. http://dx.doi.org/10.1071/MF04261. Introíni, G.O., Martins Maester, F., Pereira Leite, F.P., Recco- Pimentel, S.M., 2010. Sperm ultrastructure of Mytella (Bivalvia) populations from distinct habitats along the northern coast of S~ ao Paulo State, Brazil. Biocell 34 (3), 103e111. Jensen, J.L., Bohonak, A.J., Kelley, S.T., 2005. Isolation by distance, web service. BMC Genet. 6, 13. http://dx.doi.org/10.1186/1471-2156-6-13. Available at: http// ibdws.sdsu.edu (accessed July 2013). Keen, A.M., 1971. Sea Shells of Tropical West America. Marine Mollusks from Baja California to Peru, second ed. Stanford Unversity Press, Palo Alto, p. 1064. Kimura, M., 1981. Estimation of evolutionary distances between homologous nucleotide sequences. Proc. Natl. Acad. Sci. U. S. A. 78 (1), 454e458. Launey, S., Lepu, C., Boudry, P., Bonhomme, F., Narciri-Graven, Y., 2002. Geographic structure in the European flat oyster (Ostrea edulis F.) as revealed by microsatellite polymorphism. J. Hered. 93 (5), 331e338. ~o, J., Boudry, P., Sole -Cava, A.M., 2011. Phylogeny and phyloLazoski, C., Gusma geography of Atlantic oyster species, evolutionary history, limited genetic connectivity and isolation by distance. Mar. Ecol. Prog. Ser. 426, 197e212. http:// dx.doi.org/10.3354/meps09035. Please cite this article in press as: de Souza, T.O., et al., Population structure and identification of two matrilinear and one patrilinear mitochondrial lineages in the mussel Mytella charruana, Estuarine, Coastal and Shelf Science (2014), http://dx.doi.org/10.1016/j.ecss.2014.11.009 10 T.O. de Souza et al. / Estuarine, Coastal and Shelf Science xxx (2014) 1e10 Foighil, D., 2004. Hidden Floridian biodiversity: mitochondrial and nuclear Lee, T., O gene trees reveal four cryptic species within the scorched mussel, Brachiodontes exustus, species complex. Mol. Ecol. 13, 3527e3542. http://dx.doi.org/10.1111/ j.1365-294X.2004.02337.x. ^ncia e da capacidade de isoLeonel, R.M.V., Silva, I.N., 1988. Estudo da sobrevive lamento de Mytella guyanensis (Mollusca-Bivalvia), em diferentes salinidades. Rev. Nord. Biol. 6, 35e41. Li, W.H., 1997. Molecular Evolution. Sinauer Associates Inc., Sunderland, MA, p. 487. Luttikhuizen, P.C., Drent, J., Baker, J., 2003. Disjunct distribution of highly diverged mitochondrial lineage clade and population subdivision in a marine bivalve with pelagic larval dispersal. Mol. Ecol. 12, 2215e2229. http://dx.doi.org/ 10.1046/j.1365-294X.2003.01872.x. Murray-Jones, S.E., Ayre, D.J., 1997. High levels of gene flow in the surf bivalve Donax deltoides (Bivalvia, Donacidae) on the east coast of Australia. Mar. Biol. 128, 83e89. Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York, p. 512. -Cava, A.M., 2005. Oliveira, M.E.G.C., Russo, C.A.M., Lazoski, C., Vianna, P.R.F.G., Sole Genetic variation and population structure of two species of neo-tropical mudmussels (Mytella spp). Genet. Mol. Res. 4 (2), 197e202. , M.N., Carvalheira, J.V., 1972. Estudos preliminares sobre a ocorre ^ncia de Paranagua rio Mar. Brazil 61, 1e10. sururu na Baía de Guanabara. Inst. Pesqui. Mar. Ministe Passamonti, M., Ghiselli, F., 2009. Doubly uniparental inheritance, two mitochondrial genomes, one precious model for organelle DNA inheritance and evolution. DNA Cell. Biol. 28 (2), 79e89. http://dx.doi.org/10.1089/dna.2008.0807. ~o, M.S.N., 2003. Estimativa da Pereira, O.M., Hilberath, R.C., Ansarah, P.R.A.C., Galva rio produç~ ao de Mytella falcata e de M. guyanensis em bancos naturais do estua de Ilha Comprida e SP e Brasil. Bol. Inst. Pesca, S~ ao Paulo 29 (2), 139e149. Pleyte, K.A., Duncan, S.D., Phillips, R.B., 1992. Evolutionary relationships of the salmonid fish genus Salvelinus inferred from DNA sequences of the first Internal Transcribed Spacer (ITS 1) of ribosomal DNA. Mol. Phylogenet. Evol. 1 (3), 223e230. Posada, D., 2008. jModelTest, phylogenetic model averaging. Mol. Biol. Evol. 25 (7), 1253e1256. http://dx.doi.org/10.1093/molbev/msn083. Reece, K.S., Cordes, J.F., Stubbs, J.B., Hudson, K.L., Francis, E.A., 2008. Molecular phylogenies help resolve taxonomic confusion with Asian Crassostrea oyster species. Mar. Biol. 153, 709e721. http://dx.doi.org/10.1007/s00227.007.0846.2. ~o Universidade do Rio Rios, E.C., 1994. Seashells of Brazil, second ed. Fundaça Grande, Rio Grande, p. 492. Roessig, J.M., Woodley, C.M., Cech Jr., J.J., Hansen, L.J., 2004. Effects of global climate change on marine and estuarine fishes and fisheries. Rev. Fish Biol. Fish. 14, 251e275. Rogers, A.R., Harpending, H., 1992. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9 (3), 552e569. Rozas, J., S anchez-Del Barrio, J.C., Messeguer, X., Rozas, R., 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19 (18), 2496e2497. http://dx.doi.org/10.1093/bioinformatics/btg359. Salzburger, W., Ewing, G.B., Von Haeseler, A., 2011. The performance of phylogenetic algorithms in estimating haplotype genealogies with migration. Mol. Ecol. 20, 1952e1963. http://dx.doi.org/10.1111/j.1365-294X.2011.05066.x. Santaclara, F.J., Espineira, M., Cabado, A.G., Aldaroso, A., Gonzalez-Lavin, N., Vieites, J.M., 2006. Development of a method for the genetic identification of mussel species belonging to Mytilus, Perna, Aulacomya, and other genera. J. Agric. Food Chem. 54, 8461e8470. http://dx.doi.org/10.1021/jf061400u. Santos-Filho, C., Tagliaro, C.H., Beasley, C.R., 2008. Seasonal abundance of the shipworm Neoteredo reynei (Bivalvia, Teredinidae) in mangrove driftwood from a northern Brazilian beach. Iheringia Ser. Zool. 98 (1), 17e23. Sambrook, J., Russell, D.W., 2001. Molecular Cloning, a Laboratory Manual, third ed. Cold Spring Harbor Laboratory Press, New York, p. 2344. n larval y crecimiento del mejilo n Mytella guyanensis L. Sibaja, W.G., 1988. Fijacio (Bivalvia, Mytilidae) em Isla Chira. Costa Rica. Rev. Biol. Trop. 36 (2B), 453e456. Sirna-Terranova, M., Lo Brutto, S., Arculeo, M., Mitton, J.B., 2006. Population structure of Brachidontes pharaonis (P. Fischer, 1870) (Bivalvia, Mytilidae) in the Mediterranean Sea, and evolution of a novel mtDNA polymorphism. Mar. Biol. 159, 89e101. Sirna-Terranova, M., Lo Brutto, S., Arculeo, M., Mitton, J.B., 2007. A mitochondrial phylogeography of Brachidontes variabilis (Bivalvia: Mytilidae) reveals three cryptic species. J. Zool. Syst. Evol. Res. 45 (4), 289e298. Skibinski, D.O.F., Gallagher, C., Beynon, C.M., 1994. Mitochondrial DNA inheritance. Nature 368, 817e818. http://dx.doi.org/10.1038/368817b0. Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585e595. Tajima, F., 1993. Statistical analysis of DNA polymorphism. Jpn. J. Genet. 68, 567e595. Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S., 2011. MEGA5, molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum Parsimony methods. Mol. Biol. Evol. 28, 2731e2739. http://dx.doi.org/10.1093/molbev/msr121. Theologidis, I., Fodelianakis, S., Gaspar, M.B., Zouros, E., 2008. Doubly uniparental inheritance (DUI) of mitochondrial DNA in Donax trunculus (Bivalvia, Donacidae) and the problem of its sporadic detection in Bivalvia. Evolution 62, 959e970. http://dx.doi.org/10.1111/j.1558-5646.2008.00329.x. Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., Higgins, D.G., 1997. The CLUSTAL X windows interface, flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876e4882. Wood, A.R., Apte, S., Macavoy, E.S., Gardner, J.P.A., 2007. A molecular phylogeny of the marine mussel genus Perna (Bivalvia, Mytilidae) based on nuclear (ITS1 & 2) and mitochondrial (COI) DNA sequences. Mol. Phyl. Evol. 44, 685e698. http:// dx.doi.org/10.1016/j.ympev.2006.12.019. Yuan, W., Walters, L.J., Schneider, K.R., Hoffman, E.A., 2010. Exploring the survival threshold, A study of salinity tolerance of the nonnative mussel Mytella charruana. J. Shellfish Res. 29, 415e422. http://dx.doi.org/10.2983/035.029.0218. Zardi, G.I., Mc Quaid, C.D., Teske, P.R., Barker, N.P., 2007. Unexpected genetic structure of mussel populations in South Africa, indigenous Perna perna and invasive Mytilus galloprovincialis. Mar. Ecol. Prog. Ser. 337, 135e144. Zouros, E., 2013. Biparental inheritance through uniparental transmission, the doubly uniparental inheritance (DUI) of mitochondrial DNA. Evol. Biol. 40, 1e31. http://dx.doi.org/10.1007/s11692.012.9195.2. Please cite this article in press as: de Souza, T.O., et al., Population structure and identification of two matrilinear and one patrilinear mitochondrial lineages in the mussel Mytella charruana, Estuarine, Coastal and Shelf Science (2014), http://dx.doi.org/10.1016/j.ecss.2014.11.009