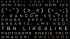RANDOM AMPLIFIED POLYMORPHIC DNA ANALYSIS OF KELAH
Transcription
RANDOM AMPLIFIED POLYMORPHIC DNA ANALYSIS OF KELAH
Second National Congress on Genetics,
l3-l5
November 1996, Genetics Society of
Malaysia
4Ss
RANDOM AMPLIFIED POLYMORPHIC DNA ANALYSIS OF KELAH (TOR TAMBROIDES)
AI\D KEJOrR ( NEOLISSOCHILUS HENDERSONI ) FISHES (CYPRINIDAE)
S.
,Ntr( MAT DAI,ID
SI]RIANIZA, ZI]LKEFLIE ZAMROD, Y, BAKAR AND
t
Department o/ Genetics and Department of Zoologt, Faculty of Lfe kiences
(Inive rsiti Kebangsaan Malaysia, B angr, Se langor
ABSTRACT
Random Amplified Polymorphic DNA (RAPD) generated using arbitrary primers of l0 nucleotide
length by Polymerase Chain Reaction (PCR) was investigated in two Cyprinidae species, kelah (?"or
tambroides) and kejor (Neolissochilus hendersoni). Electrophoresis of the amplification products
indicated that four out of the l0 primers GEN I were able to generate polymorphic products ranging in
length from 0.2-2.0 Kb. Genetic diversity index of kelah and kejor was calculated as 0.383 and 0.387,
respectively.
INTRODUCTION
Cyprinidae is ubiquitous in freshwater habitats throughout Malaysia and is the largest family in terms of the
of genome and species. Two species belonging to this family, kelah (Tor tambroides) and kejor
(Neolissochilus hendersony', which are found in the upstreams and restricted to only a few river systems are
threatened, especially by forest clearing and overfishing. They are highly prized among anglers and there is a
considerable interest to utilize these species for aquaculture. Recognizing their potential for aquaculture, a
comprehensive population study utilizing both morphometrics and molecular techniques is currently being
undertaken to assess the variability of the species for the purposes of resource conservation and management,
and exploitation for aquaculture.
number
This paper presents a preliminary results of a comparative study to determine polymorphisms among
individuals within and between kelah and kejor using Random Amplified Polymorphic DNA (RAPD)
technique. This technique is robust, simple, fast, sensitive and particularly suited to problem where specific
genetic markers are lacking or quantity of genomic DNA available is limited (Dinesh et al., 1993). A short
oligonucleotide primer of random sequence was utilized to generate polymorphic DNA fragments. Fragments
that are polymorphic at the appropriate level are used as individual, population or species-specific markers
(Bielawski et al., 1995). Therefore, RAPD markers hold the potential for considerable utility in selective
breeding, fishery management and conservation.
MATERIALS AND METHODS
Samples
Kelah and kejor were obtained from Sungai Keniam, Pahang and Sungai Pertang, Tasik Kenyir, respectively.
Ten individuals per species were utilized in the study.
Extraction of genomic DNA
DNA from individual sample was extracted following the method described by Bielawski et al. (1995) with some
436
Second National Congress on Genetics, I i-15 November 1996, Genetics Society of Malaysia
modifications. Approximately 0.0359 muscle tissue from individual fishes were cut into small pieces, suspended
and incubated with mild shaking at 37oC in 400:1 extraction buffer (lOmM Tris.Cl, pH 8.0; 2mM EDTA, pH 8.0;
10mM NaCl; l% SDS; 0.5mg/ml Proteinase K; 50mM DTT). After phenol extraction and ethanol precipitation,
DNA was dried, resuspended in sterile distilled water and stored at -20oC.
O lig onu cle o tide p r imers
set of 10 decamer primers from Genosys Biotechnologies was used in this study (Table 1). Each amplication
product is identified by its size in base pair ( bp ) following the code of each primer used in the reaction. For
example, GEN 1 60-01-3310 refers to the 3310 bp product amplified with primer GEN I 60-01. Sizes were inferred
by comparisons with a l00bp ladder (Promega).
A
Table 1: Sequence and codes of the random primers used.
Primer
codes
GEN r
GEN I
GEN l
GEN I
GEN l
GEN l
GEN I
GEN I
GEN I
GEN l
60-01
60-02
60-03
60-04
60-0s
60-06
60-07
60-08
60-09
60-10
(5' to 3')
Tm
Sequence
24.1
CGCAGTACTC
22.0
23.5
29.6
CTACACAGGC
283
24.8
2l.l
GTCCTACTCG
GTCCTTAGCG
GTCCTCAACG
CTACTACCGC
GAGTCACTCG
30.1
GTCCTCAGTG
CGTCGTTACC
21.7
GCAGACTGAG
20.4
Ampffication condition
DNA amplification were performed according to Bielawski et al (1995) with modifications. Genomic DNA was
amplified in 25:l reaction mixtures containing lX PCR buffer (Finnzymes), 2.0mM MgCl2,0.5:M decamer primer,
0.2mM of each dNTP (Promega) and I Unit of thermostable DNA Polimerase of Therrnus brockianus,
DynazymerM (Finnzymoy, Finland). Amplification was perform in a Perkin-Elmer Cetus GeneAmp PCR System
2400,programmed for 45 cycles of 30s denaturation at 94oC,30s annealing at36oC and 2min primer extention at
72oC. Samples of 25ul were analyzed by electrophoresis on l.5Yo agarose gel in 0.5X TBE (89mM Tris, pH8.3;
89mM Boric Acid; 5mM EDTA). Amplified products were detected by staining with Ethidium bromide (l:g/ml)
and visualized by illumination with ultraviolet light.
The amplifications were repeated at least twice. Only reproducible bands were used in the survey. These bands
were considered independent phenotypic markers and scored as present or absent for each individual. Chisquared analyses were used to determine if band frequencies were significantly different among species. As a
measure of genetic diversity of samples, Crow formula for genetic diversity, 1was calculated.
Ip, , ( J.F Crow, unpublished )
RESULTS AND DISCUSSION
of kelah and kejor species was investigated. The spectrum of
particular
template-primer combination. The technique of DNA
products
was reproducible for a
amplified
play
important roles in RAPD analysis. For the purpose of
amplified fragment separation and detection system
Random Amplified Polymorphic DNA (RAPD)
Second National Congress on Genetics, I3-15 November 1996, Genetics Society of Malaysia
comparison
437
of the RAPD patterns obtained, we used similar electrophoresis conditions, gel size and detection
of the
system. From the 10 primers screened, four were found to give polymorphic products based on the basis
pattern clarity and the amount of detectable polymorphisms.
Generally, 3 to 14 fragments from 3310 to 160 bp were obtained with the four primers for a total of 43 RAPD
markers. These primers gave species-specific RAPD patterns. Fig. I illustrates the profiles obtained from primer
GEN I 60-01 used to ampliff segments of genomic DNA for samples of both species. From such individual's
profiles, a matrix of presence and absence for all twenty individuals was obtained. Table 2 shows the frequencies
ior each marker for the kelah and kejor samples. Most of the markers had different frequencies between the two
species. However, eight markers were common to both species.
Figure
I
.
PCR produot for primer GEN
I
60-01
.
Table 2. Frequetrcies at 43 RAPD markers in samples of kelah and keior.
keim
Marker
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
16041-3310
kelatl
Marker
1.0
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
GEN
I 60{l-1900
0.6
0.5
0.6
0.6
0.4
I 60-01-1780
o.7
16041-1510
0.5
0.0
0.1
0.0
0.0
0.6
0.6
I 60-01-1260
16001-1200
1.0
1.0
1.0
1.0
160{l-1150
0.9
160{)14870
0.4
I 60{14720
I 5041{600
0.4
0.6
0.9
0.6
0.6
0.9
0.0
I 6041-2510
16041-2190
16041-2000
160{l{480
1604+2000
1504+1480
I
60-04-1000
16044{850
I 6044-.0600
I 60{44420
160{44350
160-044210
0.4
o.7.
1.0
o.2
0.3
0.8
0.2
0.8
0.6
0.5
0.0
0.5
0.5
0.0
1.0
1.0
0.0
keior
kelah
I 60{7-2630
0.5
5047-1900
0.4
I 60-07-1740
I 6047-1380
I 50{7-1320
0.4
0.3
0.3
0.6
o.7
0.6
0.6
0.0
0.0
0.0
0.0
0.3
1
I 60{7{190
160{7-0660
I 60{74290
16047-0160
I 6048-1380
16048-1000
160484790
1604845E0
160{8{500
160484480
160{84390
gEN 160484350
GEN 160{84320
GEN l 60-08{290
GEN 160{8{260
GEN 16048{190
0.1
1.0
1.0
1.0
1.0
l.o
1.0
0.1
0.0
0.0
0.0
0.0
0.0
0.6
0.0
1.0
0.3
1.0
1.0
1.0
1.0
1.0
1.0
0.0
0.6
o.7
0.4
0.1
438
Second National Congress on Genetics, l3-15 November 1996, Genetics Society of Malaysia
Overall there is a highly statistically significant difference in the proportion of bands in each RAPD marker in
the two species, calculated using the quick formula for Chi-squared test ( i.e. A2
= 90.69 and critical value of 12
= 55.758 atthe
5%o
significance level ).
ftelah). Eventhough the estimates do not differ befween the two species, the percentage of polymorphic markers
are different ( Table 3 ).
Table 3. Genetic diversity estimates at 43 RAPD markers *.
Population
kejor
kelah
7o
polymomhic markers Mean genetic diversity
72.09
37.21
0.387
0.383
* Only frequencies lower than 0.95 were included in the analysis.
CONCLUSION
Intrapopulation and interspecific RAPD variations was detected with 4 of l0 primers used. This preliminary RAPD
analysis is not yet sufficient in identiffing markers to separate the two species. Research work using a greater
number of primers to build up a more credible results of intra- and inter- population, and interspecific variations is
in progress, Despite the obvious limitations of the present study, nevertheless it points out the potential of this
technique to study genetic variation, as evident from this study and on other fish species (Bardakci and Skibinski,
1994 ; Allegrucci et a|.,1995).
ACKNOWLEDGEMENT
This work is supported by MPKSN grant IRPA 0l-02-02-0011
REFERENCES
Allegmrcci, G. J., Caccone, A., Cataudella, S., Powell, J.R., and Sbordoni, V. (1995). Acclimation of the European
seabass to freshwater: monitoring genetic changes by RAPD polymerase chain reaction to detect DNA
polymorphisms. Marine Biologt 121: 59i,-599.
Bardakci, F., and Skibinski, D. O. F. (1994). Application of the RAPD technique in tilapia fish: species and
subspecies identification. Heredity 1i: I l7-123.
Bielawski, J. P., Noack, K., and Pumo, D. E. (1995). Reproducible amplification of RAPD markers from vertebrata
DNA. BloZechniques 18: 856-860.
Dinesh, K. R., Lim, T. M., Chua, K. L., Chan, W. K., and Phang, V. P. E. (1993). RAPD analysis: An efficient
method of DNA fingerprinting in fishes. Zoological Science l0: 849-854.